We have found that the DNA extraction procedure of Metzenberg and Baitch (Neurospora Newsl. 28:20)/Stevens and Metzenberg (Neurospora Newsl. 29:27) while giving excellent yield and size of DNA, is somewhat cumbersome and also results in the occasional sample that proves to be uncuttable. We report here a simple method for growing Neurospora and for isolation of DNA that may be performed in two days from start to finish. The growth of mycelia in Petri plates (suggested by G. May) eliminates the need for large numbers of flasks when growing many cultures for DNA isolation.
1. Inoculate plastic Petri plates containing 25 ml of 1X Vogels, 2 sucrose, 0.04 Tween-80 (to prevent conidiation) (Zalokar, M. 1954. Arch. Biochem. Biophys. 50:71-80) plus any other required supplements. We use a loopful of conidia from a 5-10 day old tube. Incubate at 33°C. Handle the plates carefully to prevent spillage.
2. Harvest the mycelia by filtration after 36-48 h, rinse with distilled water and remove excess water by briefly pressing the pad between paper towels. Lyophilize for at least 2 h.
3. Grind the lyophilized pad in a 1.5 ml microfuge tube or a 15 ml conical tube using a metal rod or a spatula.
4. Add 1 ml of extraction buffer to the tube containing the mycelial powder and mix gently. Let stand at room temperature. for 20 min.
5. Transfer the homogenate to a microfuge tube and centrifuge in a microfuge for 5 min.
6. Recover supernatant and transfer to a tube containing 1 ml of diatomaceous earth (DE) in 6M Guanidine Thiocyanate. Let this stand at room temperature. for 2-3 min.
7. Decant into a spin column (either a 3 cc syringe body plugged with siliconized glass wool or a prefabricated spin column) and filter by aspiration. Rinse 3 times with 2 ml of 80 isopropanol. Continue aspiration for 2 min to dry out the DE. We use a standard 1 liter filter flask with a #8 stopper drilled with 5 small bore holes. Ten samples can be filtered simultaneously by using two flasks from one aspirator with a Y-junction. Commercial manifolds are also available.
8. Place each column into a 15 ml conical tube with the tip of the column resting inside a 0.5 ml microfuge tube at the bottom of the 15 ml tube. Add 150 ul of ddH20 at 65°C and spin in a tabletop centrifuge at maximum speed for 1 minute. Add an additional 150 μl and repeat.
Extraction Buffer
2M NaCl
0.4 Deoxycholic acid (sodium salt) Sigma D-6750
1.0 Brij 58 (polyoxyethylene 20 cetyl ether ) Sigma P-5884
DE/GTC
8.5 g of acid washed diatomaceous earth (Sigma D-5384), added to 100 ml of 6M Guanidine Thiocyanate (ultrapure, BMB)
Stable for approximately 6 months at room temperature.
The yield of DNA varies from 5-30 μg depending upon the amount of mycelium that is harvested. The quality of the DNA is high, with the size averaging greater than 20 kb. It digests to completion in 2 h, and has been used in Southern blots (Figure 1) and for PCR applications. The DNA is more dilute than in typical extraction procedures, but may easily be concentrated by lyophilizing or by precipitation.
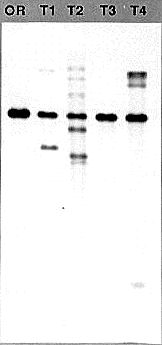
Figure 1 Genomic Southern blot analysis using DNA prepared with the Metzenberg procedure (OR, strain Oak Ridge 74-OR23-1VA) and DNAs prepared with the procedure described above (Transformants T1-T4). The visible bands range in size from >12 kb to 500 bp. One microgram of each of the DNAs was digested for 2h at 37°C with 5 units of HindIII in the manufacturers supplied buffer in a volume of 40 μl. The DNAs were fractionated overnight in a 0.7 agarose gel in 1X TAE buffer. After transfer to nylon membrane, the DNAs were probed with the transforming DNA fragment that was labeled by the random hexamer priming method. The probe is a 2.3 kb HindIII to BamHI fragment from linkage group VI, cloned from a strain where it forms a translocation junction with linkage group V just upstream of the am gene.
近年來,環狀單鏈DNA(CssDNA)因其穩定性高、免疫原性弱、可編程性強,成為基因調控、細胞治療等醫學合成生物學領域很有潛力的分子工具之一。近期,中國科學院杭州醫學研究所研究員宋杰團隊針對此前開發的......
隨著信息技術的飛速發展,傳統存儲方式已經逐漸無法滿足大數據時代的需求。在此背景下,DNA信息存儲技術應運而生,通過利用DNA分子存儲數據,已經被視為未來大規模數據存儲的潛力介質。每克DNA能夠存儲數百......
近日,我國科研人員在DNA存儲領域取得新突破,研發了一種全新的DNA存儲系統——HELIX,該系統專門用于存儲生物醫學數據,并成功實現了60MB的時空組學圖像的存儲與恢復。這一科研成果由天津大學應用數......
4月16日,深圳大學醫學部基礎醫學院、卡爾森國際腫瘤中心教授朱衛國團隊在《自然》雜志在線發表最新研究。他們揭示了連接組蛋白H1脫酰胺化修飾促進染色質開放和DNA損傷修復的機制,為腫瘤放化療的精準靶標設......
中國環境監測總站開展水生生物DNA條形碼及環境DNA分析測試公開征集工作,現向社會誠邀業界口碑良好并具有相關資質的單位參與征集。本項目資金來源:財政資金。一、項目概況:二、響應人資格要求:1.響應人須......
經過20多年的努力,科研人員成功地對6種現存猿類的基因組進行了完整測序,為研究人類進化提供了近距離視角,這被英國《自然》雜志稱為“遺傳學的一個里程碑”。123名來自多個國家和地區的科研人員組成的團隊9......
以色列耶路撒冷希伯來大學近日發布公報說,該校研究人員繪制出一份較為全面的人類基因“隱秘開關”圖譜,有助于推動遺傳疾病等方面研究。人類遺傳物質脫氧核糖核酸(DNA)上的基因可以被甲基化,這可以使相關基因......
美國芝加哥大學的科學家開發出一種名為體積DNA顯微鏡的革命性成像技術。該技術可“從內到外”繪制生命3D圖,科學家通過標記和追蹤分子間的相互作用,構建出復雜的遺傳物質3D圖,進而提供前所未有的生物體內視......
近日,中國科學院微生物研究所劉曉團隊在NucleicAcidsResearch上發表了題為“CheckpointkinasesregulatethecircadianclockafterDNAdama......
阿卜杜拉國王科技大學的一項開創性研究首次直接觀察到了DNA開始解旋的瞬間,揭示了使細胞能夠準確復制其遺傳物質的基本機制。這項研究使用冷凍電子顯微鏡和深度學習技術,捕捉到解旋酶與DNA相互作用的精微細節......